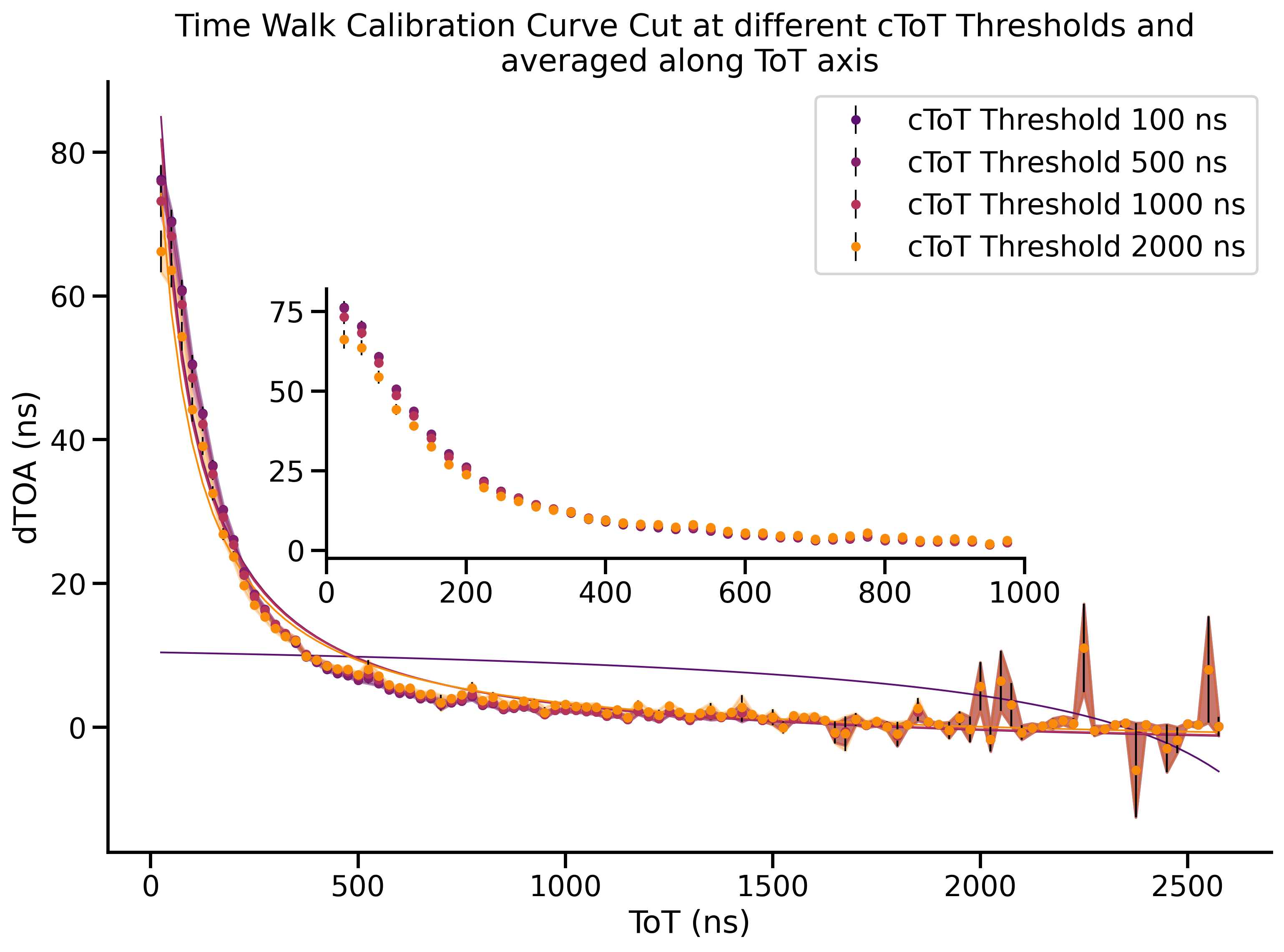
Time-walk Correction: Constructing LUT¶
While luna does this for you when you choose the ctot-cut parameter you may also compute the lookup table yourself. The method is basically computing averages of each ToT value for each cToT above a certain threshold. This threshold is the ctot-cut parameter.
In this example we also fit the lookup table to an equation that models the shape of the curve.
Note
Parameter fitting can be unstable. It is also fine to the use values as-is, without curve fitting.
Luna Command¶
./tpx3dump process -i /Users/Ciaran/atlassian-bitbucket-pipelines-runner/temp/e71169e4-520a-5b30-a5ab-ee8a44eb5fac/build/docs/source/_static/example_data.tpx3 -o /Users/Ciaran/atlassian-bitbucket-pipelines-runner/temp/e71169e4-520a-5b30-a5ab-ee8a44eb5fac/build/docs/source/_static/example_data.h5 --eps-t 170ns --eps-s 1 --calculate-timewalk-statistics --layout single
Python Script¶
Creating a lookup table from the Timewalk Matrix¶
1import os, sys
2from typing import *
3import lmfit
4import pandas as pd # ensure you have `pip install pandas`
5import warnings
6import seaborn as sns
7import matplotlib.pyplot as plt
8from matplotlib import cm
9from matplotlib.colors import Normalize
10import numpy as np
11from mpl_toolkits.axes_grid1.inset_locator import inset_axes
12
13warnings.filterwarnings("ignore") # suppress warnings from plotting libraries.
14
15sns.set_context(context="talk")
16
17# add some paths to PYTHONPATH
18for directory in ["..", "."]:
19 sys.path.append(os.path.abspath(os.path.join(os.path.dirname(__file__), directory)))
20
21# on our system "EXAMPLE_DATA_HDF5" refers to the absolute path
22# to a hdf5 file generated by luna. Replace with your own!
23from env_vars_for_docs_examples import EXAMPLE_DATA_HDF5, PLOTS_DIRECTORY
24
25# re-use functions from previous example
26from ex2_read_data_time_units import load_timewalk_matrix, TimeUnit
27
28
29def toa_calibration_equation(x, b, c, d):
30 # return b + (c / (x - d + 1e-6))
31 return b + (c / (x - d))
32
33
34def resid(params, x, y_data):
35 b = params['b'].value
36 c = params['c'].value
37 d = params['d'].value
38 y_model = toa_calibration_equation(x, b, c, d)
39 return y_model - y_data
40
41
42def fit_calibration_curve(x_data, y_data, y_data_weights=None):
43 params = lmfit.Parameters()
44 params.add('b', value=y_data[0], vary=True, min=-1e4, max=1e4)
45 params.add('c', value=1, vary=True, min=-1e4, max=1e4)
46 params.add('d', value=10, vary=True, min=-1e4, max=1e4)
47
48 model = lmfit.Model(toa_calibration_equation)
49 result = model.fit(data=y_data, params=params, x=x_data, weights=y_data_weights)
50
51 return result
52
53
54def compute_calibration_curve_data(timewalk_matrix: pd.DataFrame, cToT_thresholds: List[int]):
55 """"""
56 print(timewalk_matrix.head(5).to_string())
57 fitted_parameters_dct = {}
58 stats_data_dct = {}
59 fit_results_series = []
60 for ctot_threshold in cToT_thresholds:
61 print(f"Calculating calibration curve for ctot_threshold={ctot_threshold}")
62 # print(timewalk_matrix.head(5).to_string())
63 data_subset_df = timewalk_matrix[timewalk_matrix["CToT"] >= ctot_threshold]
64 stats_df = data_subset_df.groupby("ToT").agg(["mean", "std", "sem", "count"])["AverageDToA"]
65 stats_df = stats_df[stats_df['count'] >= 10]
66 stats_data_dct[ctot_threshold] = stats_df
67 x_data = stats_df.index.values
68 y_data = stats_df["mean"].values
69 fit_result = fit_calibration_curve(x_data=x_data, y_data=y_data, y_data_weights=None)
70 fit_results_series.append(pd.Series(fit_result, index=[ctot_threshold]))
71
72 fitted_parameters_dct[ctot_threshold] = fit_result.values
73 fitted_parameters = pd.DataFrame.from_dict(fitted_parameters_dct)
74 fit_result_df = pd.concat(fit_results_series)
75 stats_data = pd.concat(stats_data_dct)
76 stats_data.index.names = ["ctot_threshold", "ToT"]
77 return fitted_parameters, stats_data, fit_result_df
78
79
80def plot_calibration_curve(stats_data: pd.DataFrame, fit_result: pd.DataFrame):
81 # Create a colormap for shades of green
82 cmap = cm.inferno
83 # Normalize the colormap based on the cToT_thresholds
84 cToT_thresholds = list(set(stats_data.index.get_level_values(0)))
85 colours_normed = Normalize(vmin=min(cToT_thresholds) - 1000, vmax=max(cToT_thresholds) + 1000)
86
87 # Create the plot
88 fig, ax = plt.subplots(figsize=(12, 8))
89 sns.despine(fig=fig)
90
91 for ctot_threshold, df in stats_data.groupby(by="ctot_threshold"):
92 color = cmap(colours_normed(ctot_threshold))
93 # Plot the experimental data
94 x_data = stats_data.loc[ctot_threshold].index.values
95 y_data_experimental = stats_data.loc[ctot_threshold, "mean"]
96 y_err = stats_data.loc[ctot_threshold, "sem"]
97 y_data_fitted = fit_result[ctot_threshold].best_fit
98 ax.fill_between(x_data, y_data_experimental - y_err, y_data_experimental + y_err, color=color, alpha=0.3)
99 # Plot markers and fitted line
100 ax.plot(x_data, y_data_fitted, color=color, linewidth=1)
101
102 ax.errorbar(x_data, y_data_experimental, yerr=y_err, label=f'cToT Threshold {ctot_threshold} ns', marker=".",
103 color=color, linestyle="None", elinewidth=1, ecolor="black")
104
105 # Add title and labels
106 ax.set_title('Time Walk Calibration Curve Cut at different cToT Thresholds and \naveraged along ToT axis')
107 ax.set_xlabel('ToT (ns)')
108 ax.set_ylabel('dTOA (ns)')
109 ax.legend(loc='upper right', bbox_to_anchor=(1.0, 1.0))
110
111 # Create an inset Axes instance at a given position with a given size
112 # The first two arguments are the lower left corner position of the inset axes in the parent axes
113 # The next two arguments are the width and height of the inset axes
114 # bbox_to_anchor can be used to position the inset relative to the parent axes
115 ax_inset = inset_axes(ax, width="60%", height="70%", loc='upper right', bbox_to_anchor=(-0.2, 0.25, 1.0, 0.5),
116 bbox_transform=ax.transAxes)
117 sns.despine(ax=ax_inset)
118
119 # Plot the same data on the inset but for the x-axis from 0 to 1000
120 for ctot_threshold, df in stats_data.groupby(by="ctot_threshold"):
121 color = cmap(colours_normed(ctot_threshold))
122 x_data = df.index.get_level_values(1) # Or your specific way to get x_data
123 y_data_experimental = df["mean"]
124 y_data_err = df["sem"]
125 y_data_fitted = fit_result[ctot_threshold].best_fit
126
127 # Filter data for x_data < 1000
128 mask = x_data < 1000
129 x_data_inset = x_data[mask]
130 y_data_experimental_inset = y_data_experimental[mask]
131 y_data_fitted_inset = y_data_fitted[mask]
132 y_err_inset = y_data_err[mask]
133
134 # ax_inset.plot(x_data_inset, y_data_experimental_inset, marker=".", color=color, linestyle="None", alpha=0.7)
135 ax_inset.errorbar(x_data_inset, y_data_experimental_inset, yerr=y_err_inset,
136 label=f'cToT Threshold {ctot_threshold} ns', marker=".",
137 color=color, linestyle="None", elinewidth=1, ecolor="black")
138
139 # Set the inset's x-axis limit
140 ax_inset.set_xlim(0, 1000)
141
142 fname = os.path.join(PLOTS_DIRECTORY, f"ex9_timewalk_lut_construction.png")
143 plt.savefig(fname, dpi=300, bbox_inches='tight')
144
145
146if __name__ == "__main__":
147 toa_unit = TimeUnit.Nanoseconds
148
149
150 timewalk_matrix = load_timewalk_matrix(EXAMPLE_DATA_HDF5, toa_unit=toa_unit)
151
152 if timewalk_matrix is not None:
153 tot_thresholds = [100, 500, 1000, 2000]
154 params, data, fit_result = compute_calibration_curve_data(timewalk_matrix, tot_thresholds)
155 plot_calibration_curve(data, fit_result)
Script Output¶
Example Output¶
<KeysViewHDF5 ['Clusters', 'ExposureTimeBoundaries', 'PixelHits', 'TimewalkLookupTable', 'TimewalkMatrix']>
CToT ToT AverageDToA SumSquareDiff Count Std Sem
0 450 25 96.223969 53196212.0 12 21.054733 6.077978
1 450 50 83.750008 119628920.0 25 21.875000 4.375000
2 450 75 76.627617 66283176.0 24 16.618660 3.392270
3 450 100 62.467461 91039528.0 48 13.771915 1.987805
4 450 125 51.258690 118634712.0 72 12.836293 1.512772
Calculating calibration curve for ctot_threshold=100
Calculating calibration curve for ctot_threshold=500
Calculating calibration curve for ctot_threshold=1000
Calculating calibration curve for ctot_threshold=2000